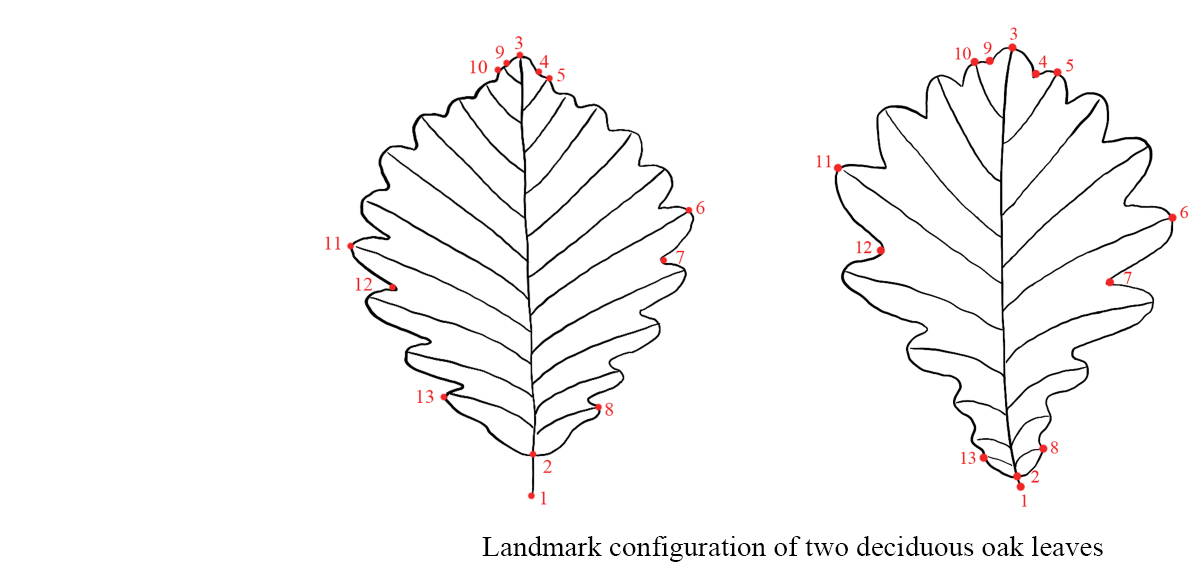
Leaf shape is extraordinary complex in oaks. The great variation of leaf morphology and the difficulty of species identification in oaks have attracted the attention of evolutionary biologists since Charles Darwin, in The Origin of Species (1872) he argued that additional studies of oaks in Britain had not resulted in greater taxonomic clarity but rather in greater confusion. Luckily, recent advance in discrimination technology provides opportunities for us to understand the leaf morphology variation in oaks. Here we will introduce our ongoing leaf morphological study in Chinese oaks by geometric morphometric methods (GMMs) and deep learning. With landmark-based geometric morphometric approach, all aspects of shape variation among a set of landmarks are captured, so specific changes can be detected without having to be specified and explicitly measured a priori. Using GMMs we had demonstrated that two congeneric deciduous oak species in sympatric areas were efficiently discriminated (Liu et al., 2018; Du et al., 2022). Deep learning promises the ability to help feature extraction quickly and effectively of the complex datasets such as numerous leaf image. The convolutional neural network (CNN) Xception architecture was used to classify the deciduous Chinese oaks and their accuracy and efficiency was compared with other traditional discrimination methods (Qi et al., 2024).
We have summarized the leaf morphology workflow and published it on this website: https://www.oakofchina.org/wp-content/uploads/2025/04/leaf-morphology-workflow-3-.pdf.
Besides used for taxonomic purposes, leaf shape is also with great functional significance, including in oaks. By multivariate analysis of leaf shape, climate factors and neutral genetic data in the alpine oak, Quercus aquifolioides, we demonstrated that leaf morphological variation across its geographic range is related to both its genetic differentiation and, to a lesser extent, to climatic factors. We discuss how these patterns could be interpreted in terms of both geographical isolations among and within lineages, and possible adaptive responses for particular traits (Li et al., 2021).
In this website we display four leaf images for each Chinese oak species in the “leaf scan image” section and we list the leaf image samples stored in our lab in BJFU, please contact us for more information.
| Species | # Populations | #Trees | #Leaves | Genetic data |
| Q. aliena | 22 | 149 | 745 | 1 |
| Q. dentata | 32 | 322 | 1610 | 1 |
| Q. serrata | 30 (3 pops from Japan) | 248 | 1195 | 1 |
| Q. fabri | 23 | 186 | 930 | 1 |
| Q. mongolica | 4 | 35 | 170 | 1 |
| Q. griffithii | 8 | 69 | 345 | 1 |
| Q. malacotricha | 2 | 11 | 55 | 1 |
| Q. aquifolioides | 29 | 170 | 850 | 3, 4, 5, 6 |
| Q. spinosa | 43 | 376 | 1881 | 3, 5 |
| Q. guyavaefolia | 6 | 45 | 225 | 2, 5 |
| Q. semecarpifolia | 5 | 31 | 154 | 2, 5 |
| Q. rehderiana | 9 | 58 | 290 | 2, 5 |
| Q. senescens | 7 | 55 | 275 | 2, 5 |
| Q. monimotricha | 7 | 51 | 254 | 2, 5 |
Note:
1: SSR 12 loci;
2: SSR 15 loci;
3: SSR 25 loci;
4: four cpDNA fragments (trnH-psbA, rpS16, trnS–trnT and trnQ–trnS);
5: SNPs from 30× resequencing for part of the individuals;
6: SNPs candidate gene of drought.
The detail for the species image from field please see section Species.
Last update: November of 2023
Participants
This work is collaborated with Prof. JijunTang (USC, USA) and Prof. Fei Guo (Tianjin University, China) for deep learning analysis;
Dr. Yuanye Zhang (Xiameng University, China); Dr. Saneyoshi Ueno (FFPRI, Japan) and Dr. Pauline Garnier-Géré (INRAE, France) for morphology analysis.
We thank graduate and undergraduate students from BFU for image scan, landmark and morphology data analysis, they are: master student: Jia Song (2013-2016); Yuan Liu (2015-2018); Yuanjuan Li (2017-2020); Wei Su (2018-2021); Yi Zhang (2020-), undergraduate student: Xiaojing Wang (2014-2018); Xuanying Wang (2016-2020); Panpan Wang (2017-2021) and PhD candidate: Min Qi (2021-).
